PMI for images (scratch)
Posted: 2016-08-01 , Modified: 2016-09-05
Tags: PMI, neural nets, vision
Posted: 2016-08-01 , Modified: 2016-09-05
Tags: PMI, neural nets, vision
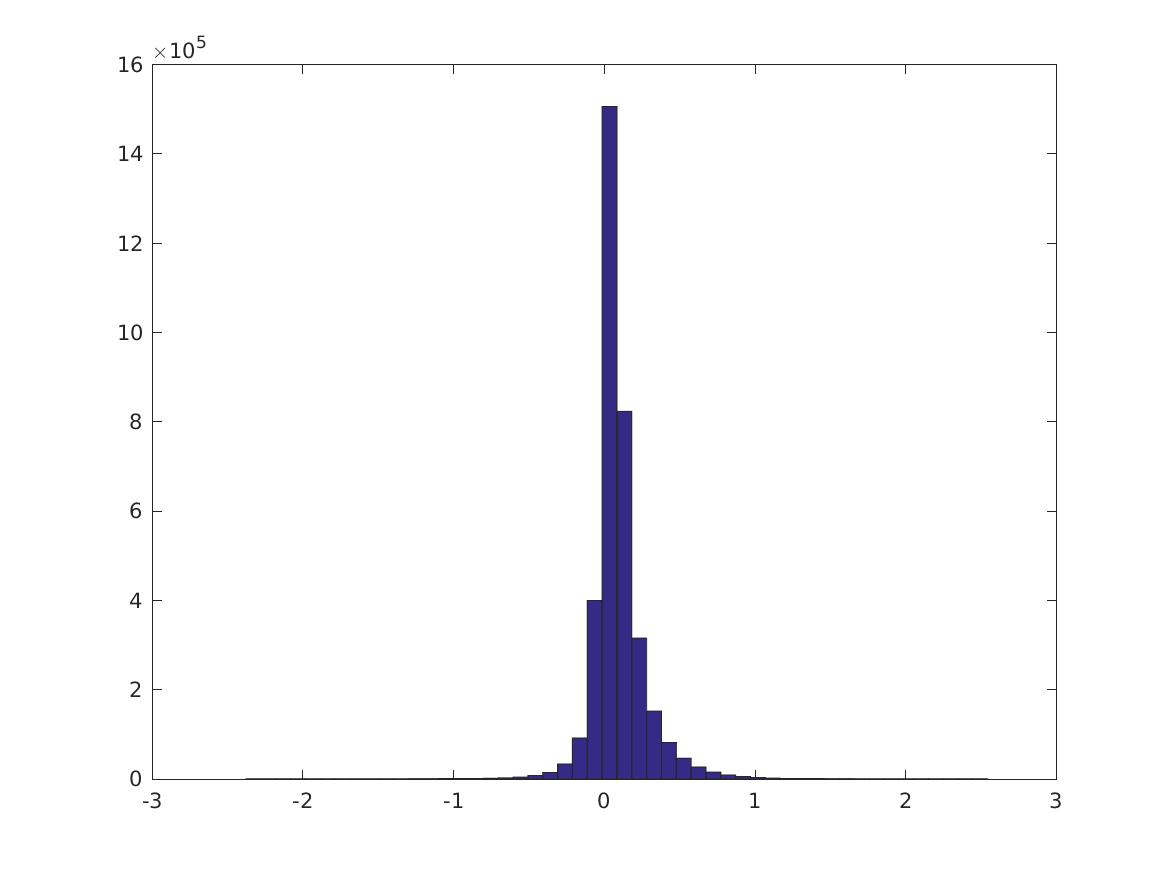
Over all pairs of features \(i,j\), this is the distribution of \(PMI(v_i,v_j)\):
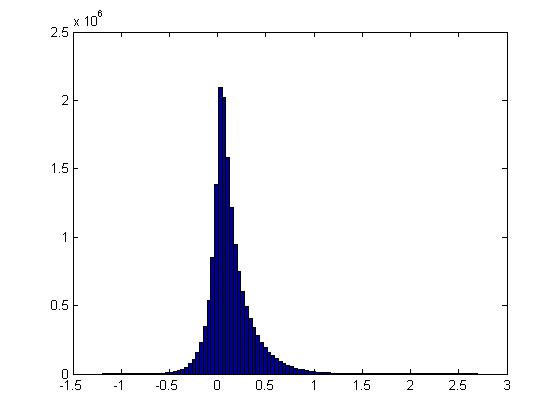
(Question: is this what you would expect if the distribution were “random” or are the tails different than what we would expect?)
Is there a correlation between PMI and distance between features? (Distance ranges from \(0\) to \(5\sqrt 2\).) It seems not.
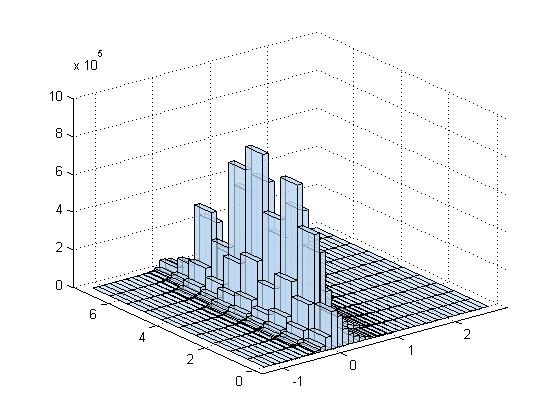
Conditioned on the digit being a specific value, how does the PMI distribution look? Still about the same… except that 1 has much larger activation. These are digits 0-4.
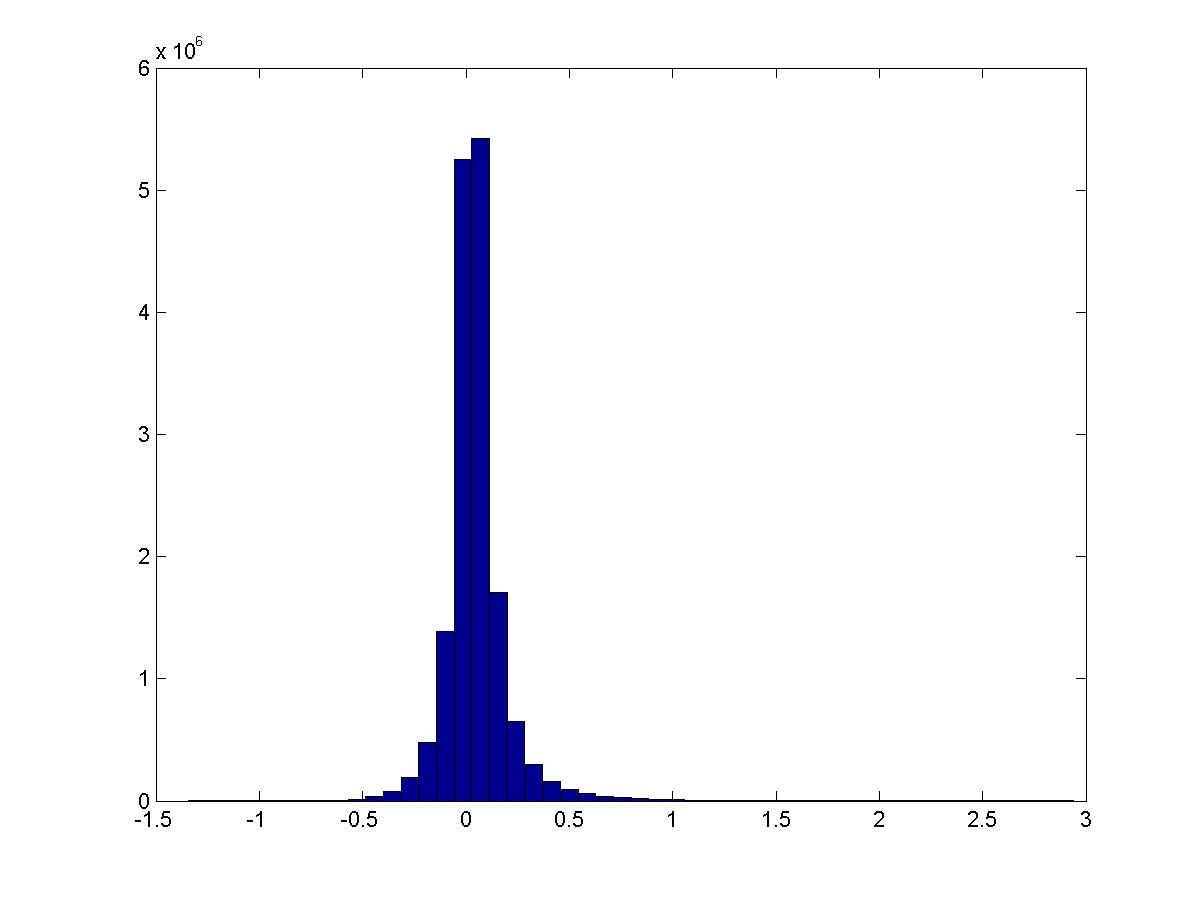
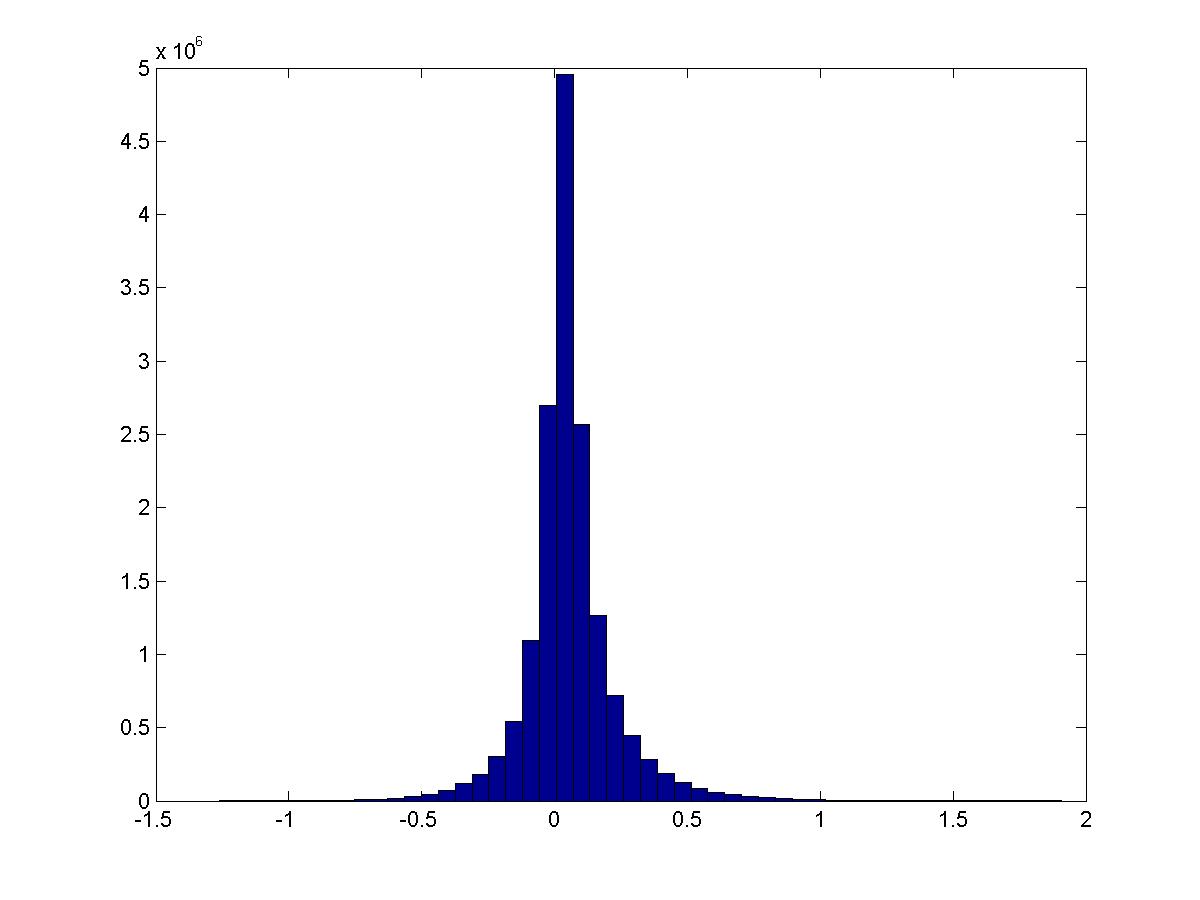
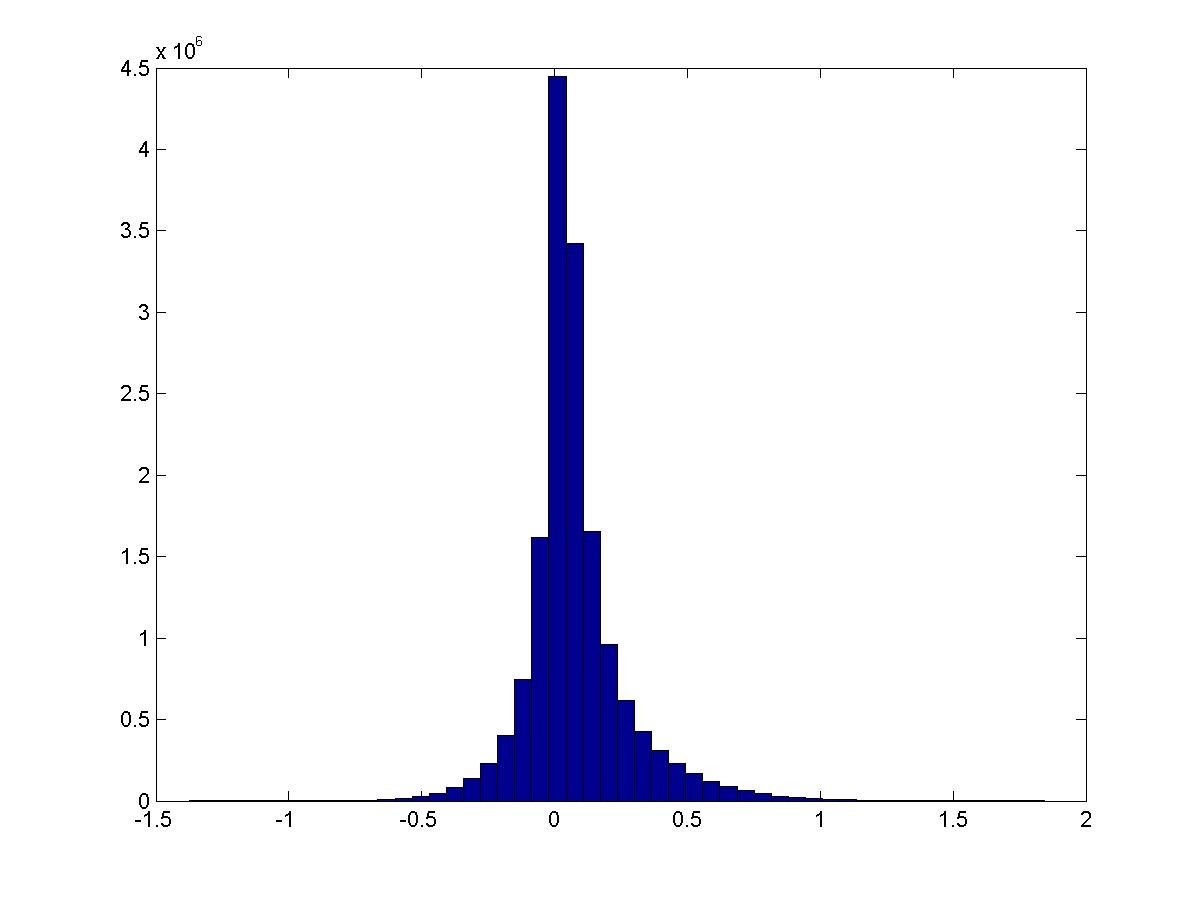
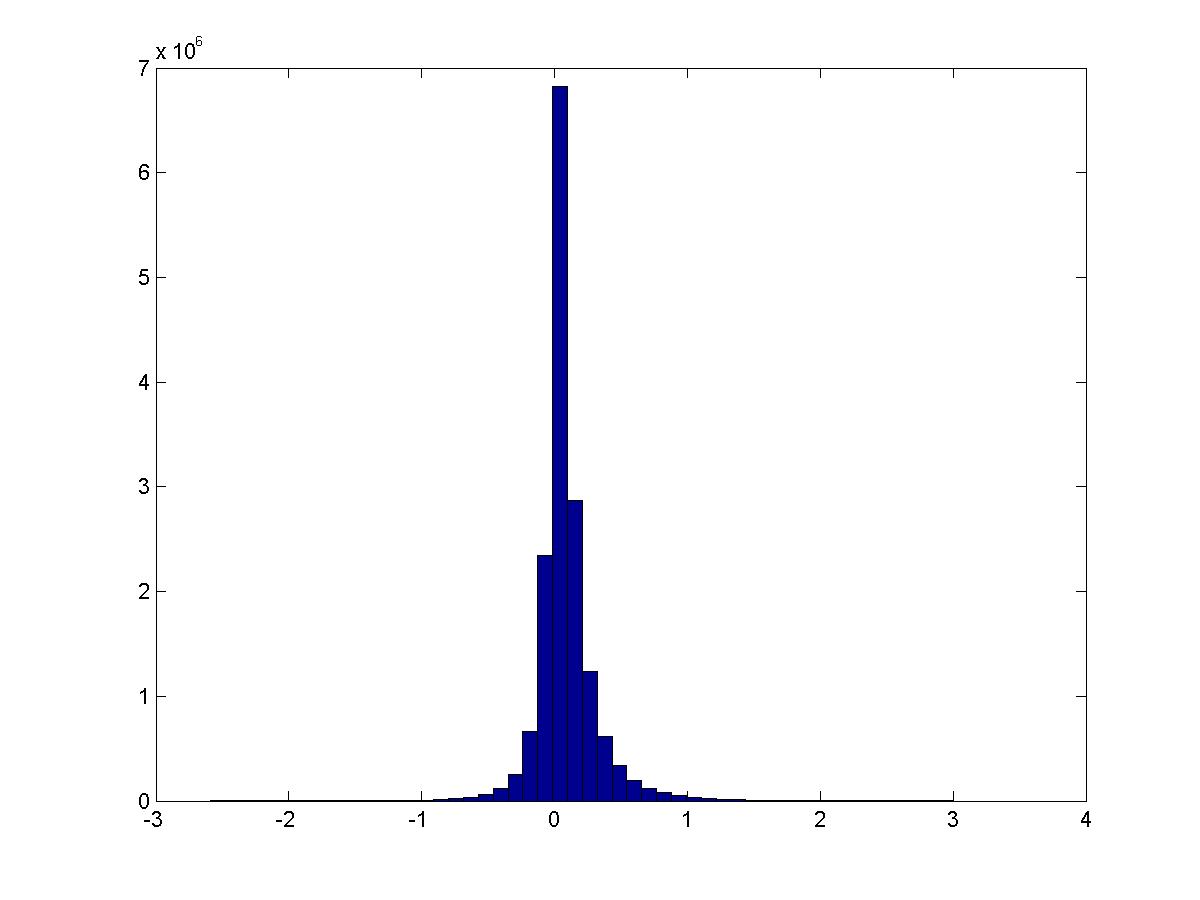
See script pmi_thresholded.m.
Remove all features that do not have PMI \(>1\) with another feature. Now do the same thing.
This is the distribution of \(PMI(v_i,v_j)\):

These are digits 0-4.




Percentiles of the distributions (\(i\)th row is 0, 10, 20, … 100% for the CPMI’s of digit \(i\), last row is PMI over all digits).
Columns 1 through 7
-0.9746 -0.0588 -0.0140 0.0112 0.0306 0.0488 0.0681
-3.1365 -0.3102 -0.0940 0.0050 0.0609 0.1144 0.1812
-0.9300 -0.0626 -0.0130 0.0134 0.0333 0.0526 0.0740
-0.7660 -0.0462 -0.0022 0.0210 0.0403 0.0602 0.0835
-2.0740 -0.0565 -0.0071 0.0188 0.0397 0.0611 0.0855
-0.8674 -0.0628 -0.0077 0.0229 0.0471 0.0705 0.0966
-2.3770 -0.0457 -0.0007 0.0228 0.0428 0.0639 0.0886
-2.4824 -0.0928 -0.0235 0.0121 0.0379 0.0633 0.0925
-0.9435 -0.0521 -0.0066 0.0161 0.0347 0.0545 0.0785
-2.1549 -0.0505 -0.0020 0.0231 0.0446 0.0679 0.0961
-0.8693 -0.0515 0.0008 0.0344 0.0640 0.0949 0.1309
Columns 8 through 11
0.0910 0.1215 0.1730 1.5181
0.2772 0.4314 0.7470 5.3609
0.1007 0.1393 0.2117 1.5297
0.1141 0.1604 0.2521 1.6011
0.1169 0.1635 0.2537 2.2486
0.1293 0.1773 0.2773 2.0363
0.1215 0.1727 0.2771 2.5443
0.1305 0.1882 0.3019 2.9006
0.1111 0.1639 0.2788 2.1102
0.1344 0.1957 0.3210 2.7701
0.1761 0.2406 0.3557 1.7908
See script pmi_thresholded.m.
Remove all features that do not have PMI \(>1\) with another feature. Now do the same thing.
This is the distribution of \(PMI(v_i,v_j)\):

These are digits 0-4.





CODE: Run pmi_thresholded and check_top_pmis.
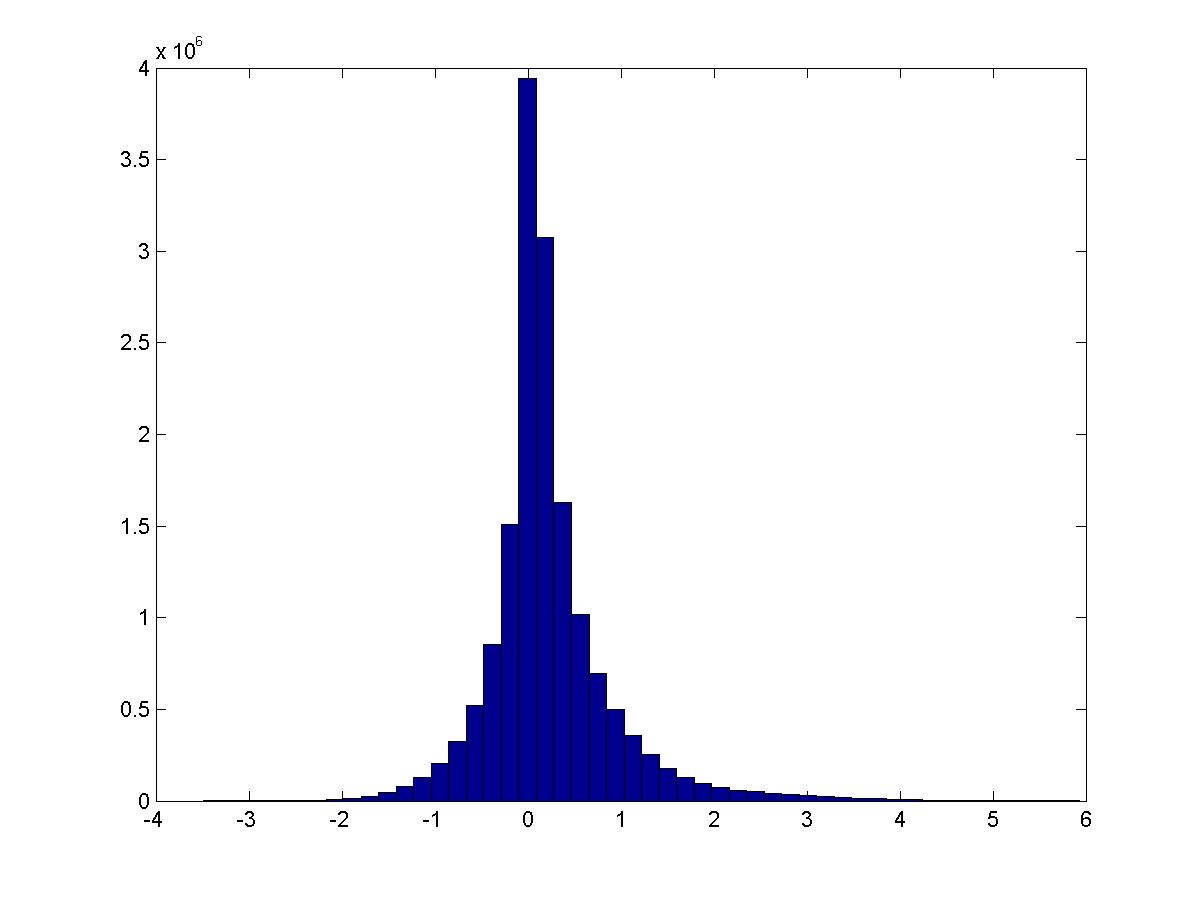
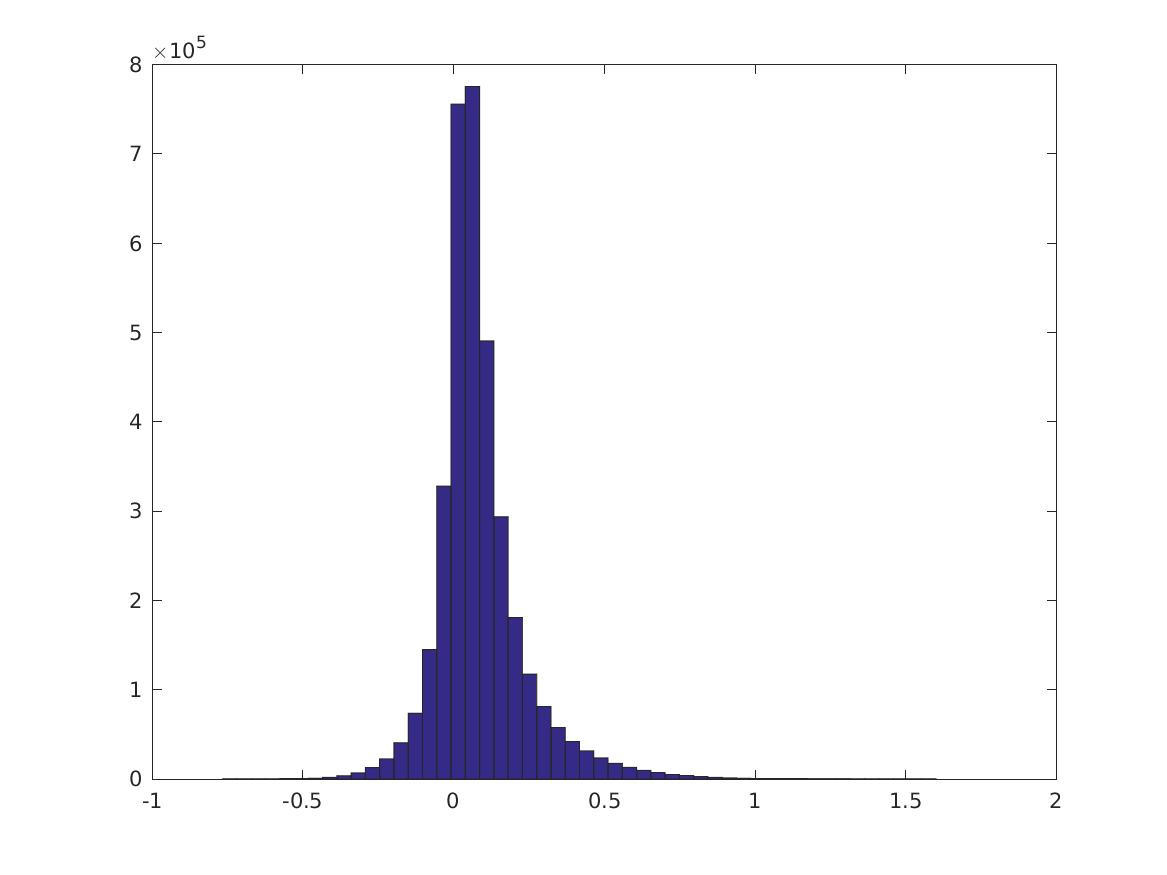
Remove all features that do not have activation \(>1\) in some image. Now do the same thing. (NOTE: the maximum PMI is less because the max PMI’s are between features that have small activation. Try this again thresholding by PMI?)
This is the distribution of \(PMI(v_i,v_j)\):

These are digits 0-9.









Percentiles of the distributions (\(i\)th row is 0, 10, 20, … 100% for the CPMI’s of digit \(i\), last row is PMI over all digits). (Note the last column is different)
Columns 1 through 7
-0.9746 -0.0588 -0.0140 0.0112 0.0306 0.0488 0.0681
-3.1365 -0.3102 -0.0940 0.0050 0.0609 0.1144 0.1812
-0.9300 -0.0626 -0.0130 0.0134 0.0333 0.0526 0.0740
-0.7660 -0.0462 -0.0022 0.0210 0.0403 0.0602 0.0835
-2.0740 -0.0565 -0.0071 0.0188 0.0397 0.0611 0.0855
-0.8674 -0.0628 -0.0077 0.0229 0.0471 0.0705 0.0966
-2.3770 -0.0457 -0.0007 0.0228 0.0428 0.0639 0.0886
-2.4824 -0.0928 -0.0235 0.0121 0.0379 0.0633 0.0925
-0.9435 -0.0521 -0.0066 0.0161 0.0347 0.0545 0.0785
-2.1549 -0.0505 -0.0020 0.0231 0.0446 0.0679 0.0961
-0.8693 -0.0515 0.0008 0.0344 0.0640 0.0949 0.1309
Columns 8 through 11
0.0910 0.1215 0.1730 1.5181
0.2772 0.4314 0.7470 5.3609
0.1007 0.1393 0.2117 1.5297
0.1141 0.1604 0.2521 1.6011
0.1169 0.1635 0.2537 2.2486
0.1293 0.1773 0.2773 2.0363
0.1215 0.1727 0.2771 2.5443
0.1305 0.1882 0.3019 2.9006
0.1111 0.1639 0.2788 2.1102
0.1344 0.1957 0.3210 2.7701
0.1761 0.2406 0.3557 1.7908
TODO: add data from check_top_pmis. This produces a graph showing which features are close to each other.
Run qsub svd_svm.cmd which runs svd_testing2.m, or sbatch slurm_svd.cmd. The results are saved in accs_1.mat.
Results:
| Dimension | Last | Best |
|---|---|---|
| 500 | 99.26 | 99.44 |
| 100 | 97.5 | 98.82 |
| 50 | 96.84 | 96.84 |
Do weighted SVD (as in the NLP paper) and then train a SVM. Note this does worse than just SVD! Why?
| Dimension | Last | Best |
|---|---|---|
| 50 | 95.44 | 96.8 |
Is PMI the right measure? Are the feature vectors close to sparse? if the average image there is \(>20\%\) activation, then no.
For the first 10 images, this is what the percentiles look like:
percentiles:
pcs =
100 99 95 90 80 70
Image 1 (label 5)
2.3230
1.2966
0.8840
0.6766
0.4381
0.3140
Image 2 (label 0)
2.1275
1.3604
0.9482
0.7418
0.5036
0.3769
Image 3 (label 4)
1.7975
0.9874
0.6185
0.4704
0.3145
0.2247
Image 4 (label 1)
2.2506
1.2399
0.7160
0.4949
0.3029
0.1992
Image 5 (label 9)
2.0837
1.3970
0.8726
0.6461
0.3937
0.2707
Image 6 (label 2)
2.4831
1.4978
1.0466
0.7880
0.5002
0.3544
Image 7 (label 1)
2.5064
1.3688
0.7591
0.5203
0.3089
0.1935
Image 8 (label 3)
3.3030
1.6943
1.1588
0.8658
0.5756
0.4070
Image 9 (label 1)
2.2263
0.9396
0.4905
0.3279
0.1944
0.1178
Image 10 (label 4)
2.2420
1.3486
0.8494
0.6169
0.3751
0.2586
This looks pretty good—the top 1% of features are much larger than the rest.
Look for discriminative features, those that occur with one label and rarely for others. (I arbitrarily define this as: the maximum average activation is \(>1.5\) times the second largest, and the maximum is at least 0.1. I exclude the digit 1 (indexed as 2).)
sig_act_inds = intersect(find(m1>=1.5*m2), find(m1>0.1));
sig_act_inds2 = intersect(sig_act_inds, find(am1 ~=2));115/7200 features fit this criterion.
Do SVM coefficients have negative weights?
collect, same patch; no collect, same patch; horiz collect; horiz no collect; vert collect; vert no collect; all
ans =
Columns 1 through 7
-0.9605 -0.3457 -0.2067 -0.1000 -0.0103 0.0783 0.1698
-1.0872 -0.0690 -0.0062 0.0281 0.0563 0.0880 0.1277
-1.5676 -0.3858 -0.2386 -0.1310 -0.0454 0.0409 0.1303
-0.9235 -0.0241 0.0307 0.0621 0.0919 0.1266 0.1695
-1.3763 -0.3810 -0.2237 -0.1053 -0.0072 0.0839 0.1708
-0.9163 -0.0108 0.0284 0.0530 0.0796 0.1093 0.1458
-1.1984 -0.0727 -0.0094 0.0253 0.0534 0.0847 0.1241
Columns 8 through 11
0.2735 0.3922 0.5678 1.3093
0.1821 0.2624 0.4080 2.4518
0.2220 0.3257 0.4557 1.1007
0.2240 0.3054 0.4534 2.6332
0.2607 0.3593 0.4863 0.9118
0.1942 0.2660 0.3970 2.6332
0.1778 0.2587 0.4034 2.6897I sorted pairs of features by PMI value and took the largest 10000 pairs. The majority of the pairs (~77%) have strongest activation with the digit 1. (The digit 1 has conditional PMI values that are much larger than all the other digits - don’t know what to do about this.) 85% of the pairs have highest activations with the same digit.
If the pair has strong activation with 1, then it’s often much larger than the other activations.
Otherwise this is not necessarily true. A typical non-1 feature has average activations with the 10 digits that looks something like this:
Columns 1 through 7
0.0708 0.2673 0.0940 0.1537 0.2122 0.1116 0.2627
Columns 8 through 10
0.2947 0.1448 0.3361
Experiments:
mnist_testing.m: Prep data.svd_testing.m: How does reducing the number of features through SVD affect classification accuracy?pairwise_mi.m: Tests related to mutual informationmi.mMI_GG.mrsvd.m nlayers: 2
layer: {2x1 cell}
type_zerolayer: 2
ndesc: 7200
numlayer: 1
centering: 0
median_contrast_normalization: 0
npatch: 5
subsampling: 2
smap: 50
type_zerolayer: 2
sigma: 0.7459
Z: [25x50 double]
w: [50x1 double]In pairwise_mi.m there’s no normalization by scaling (\(-2\log\pat{scale}\)).
https://en.wikipedia.org/wiki/Conditional_mutual_information
The right PMI measure to use here is \[ \ln \pf{\an{v,w}}{\an{v,\one}\an{w,\one}}. \] Because if we assume the activations are like \((e^{\chi,v})_\chi\), then we still get that the expected dot product of these is \(\int e^{\an{\chi,v}}e^{\an{\chi,w}}\,d\chi\propto \exp\pa{\fc{\ve{v+w}^2}{2}}\).
Do weighted SVD on PMI matrix.
Geometry
(Compare to purely random.)
Understand loss function for PMI.
Qs
nrm=mean(sqrt(sum(compTr.^2)))?Run experiments.
nohup nice -19 matlab -nodisplay -nodesktop -nojvm -nosplash < run_experiments2.m > output_2016-08-07.txt 2>&1 &%Get psiTr
pmi_experiments('logs/model_mnist_5_2_0_2_2_0_50_200_0_1.000000e-01_1.000000e-01_1.000000e-01_2.mat');
%get percentiles for each feature
pc= get_percentiles(psiTr);
save('pc_1.mat','pc','-v7.3');
%calculate PMI after normalizing and deleting zero rows.
[P, inds] = calculate_pmi(psiTr); %inds give the indices that are kept (indices of nonzero rows)
%exploratory analysis
%pmi_experiments(P);
[all_pmi_pairs, positions, dists] = pmi_experiments2(P, inds');
save('all_pmi_pairs_1.mat','all_pmi_pairs');
save('positions_1.mat','positions');
save('dists_1.mat','dists');
%cpmi matrices
Ps = calculate_cpmi(psiTr, Ytr, '1');
cpmi_experiments('P_1');psiTr_1.mat, psiTe_1.mat etc. are the feature vectors for the training/test sets and the different architectures. The architectures are
test_ckn([5 2 0],[2 2 0],[50 200 0],2,'mnist'); (7200, 0.57%)test_ckn([1 3 0],[2 2 0],[12 50 0],3,'mnist'); (1800, 0.63%)test_ckn([1 3 0],[2 4 0],[12 400 0],3,'mnist'); (3600, 0.41%)