PMI for images
Posted: 2016-08-01 , Modified: 2016-09-15
Tags: PMI, neural nets, vision
Posted: 2016-08-01 , Modified: 2016-09-15
Tags: PMI, neural nets, vision
CODE: First calculate psiTr. (It has been saved as data/psiTr_1.mat.) Then run calculate_cpmi_unnormalized_script.
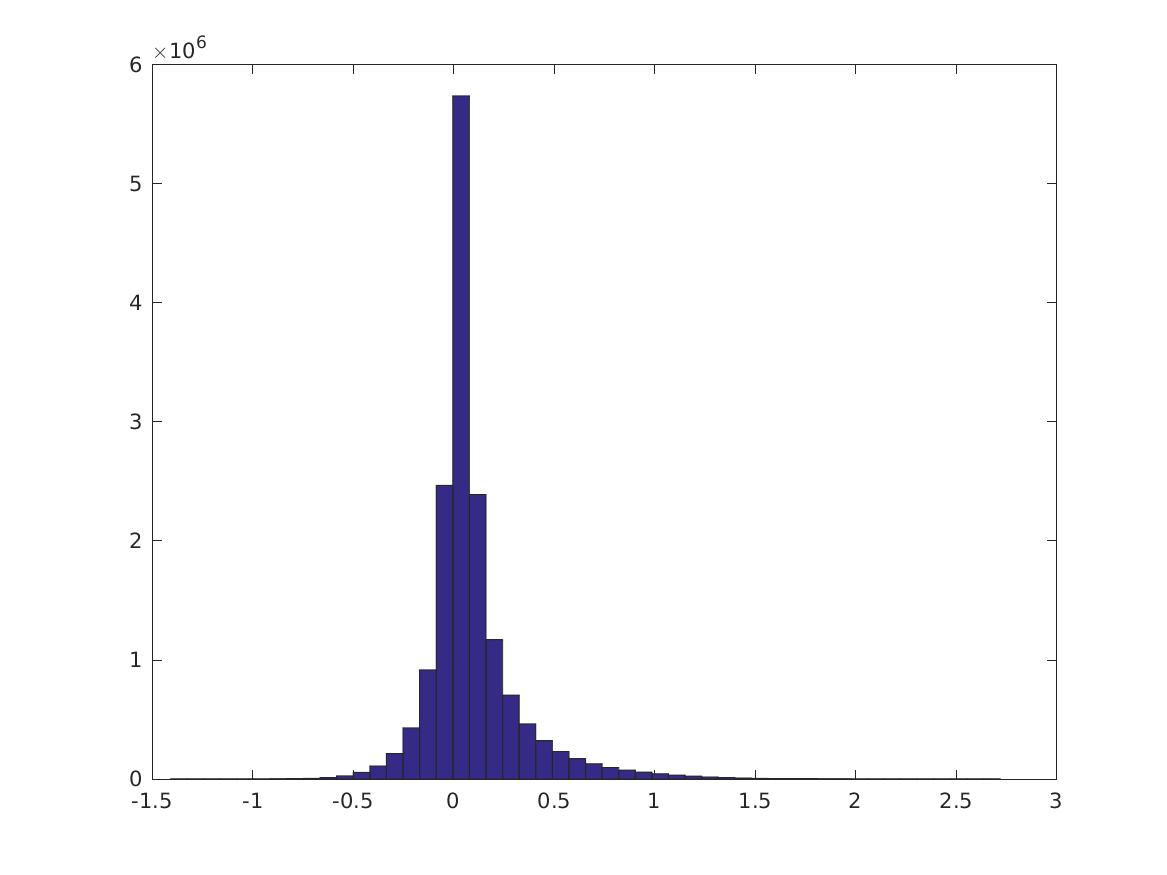
Over all pairs of nonzero features \(\{i,j\}\), this is the distribution of \(PMI(v_i,v_j)\). Note the long tails. Here we don’t normalize the features first. (You can also try with various normalizations, by passing in a normalization function to calculate_cpmi_unnormalized.) There are 5652/7200 nonzero features.

Conditioned on the digit being a specific value, how does the PMI distribution look? Still about the same… except that 1 has much larger activation. These are digits 0-9 in order.
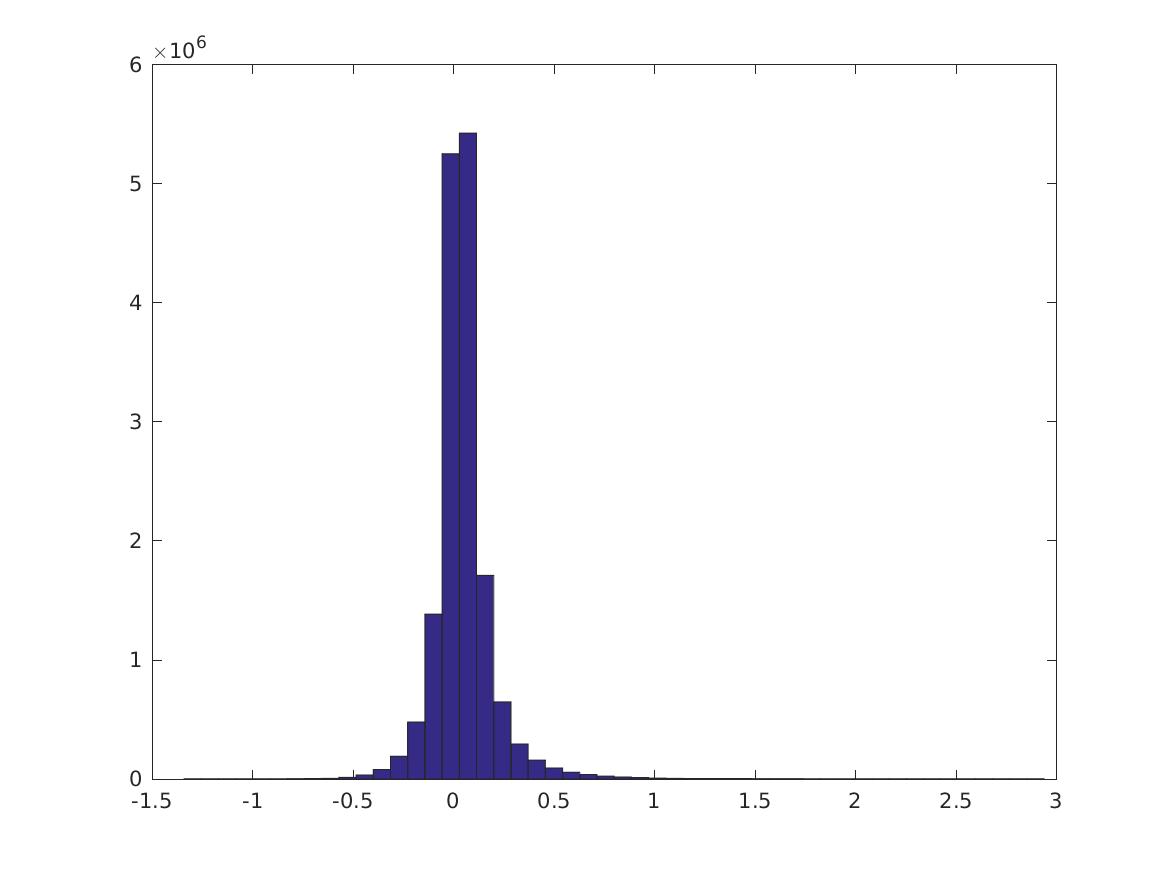



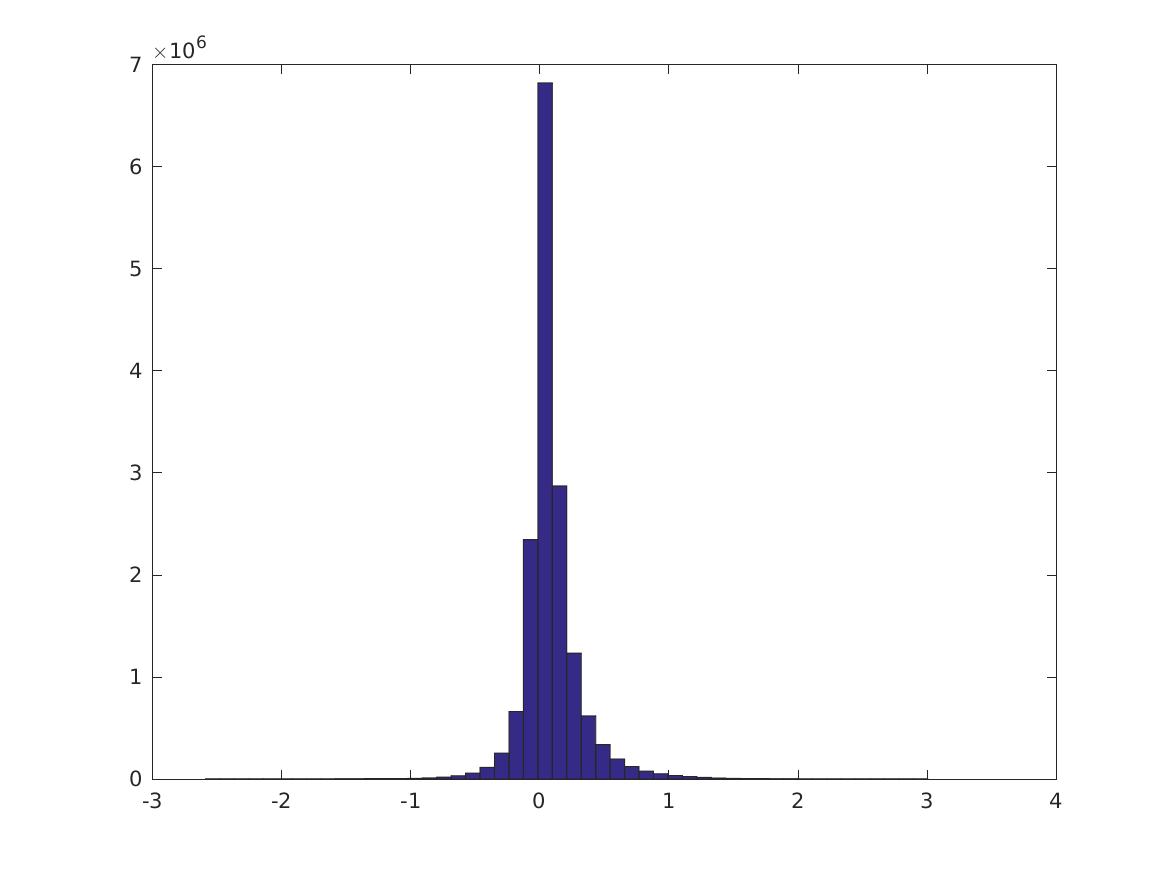
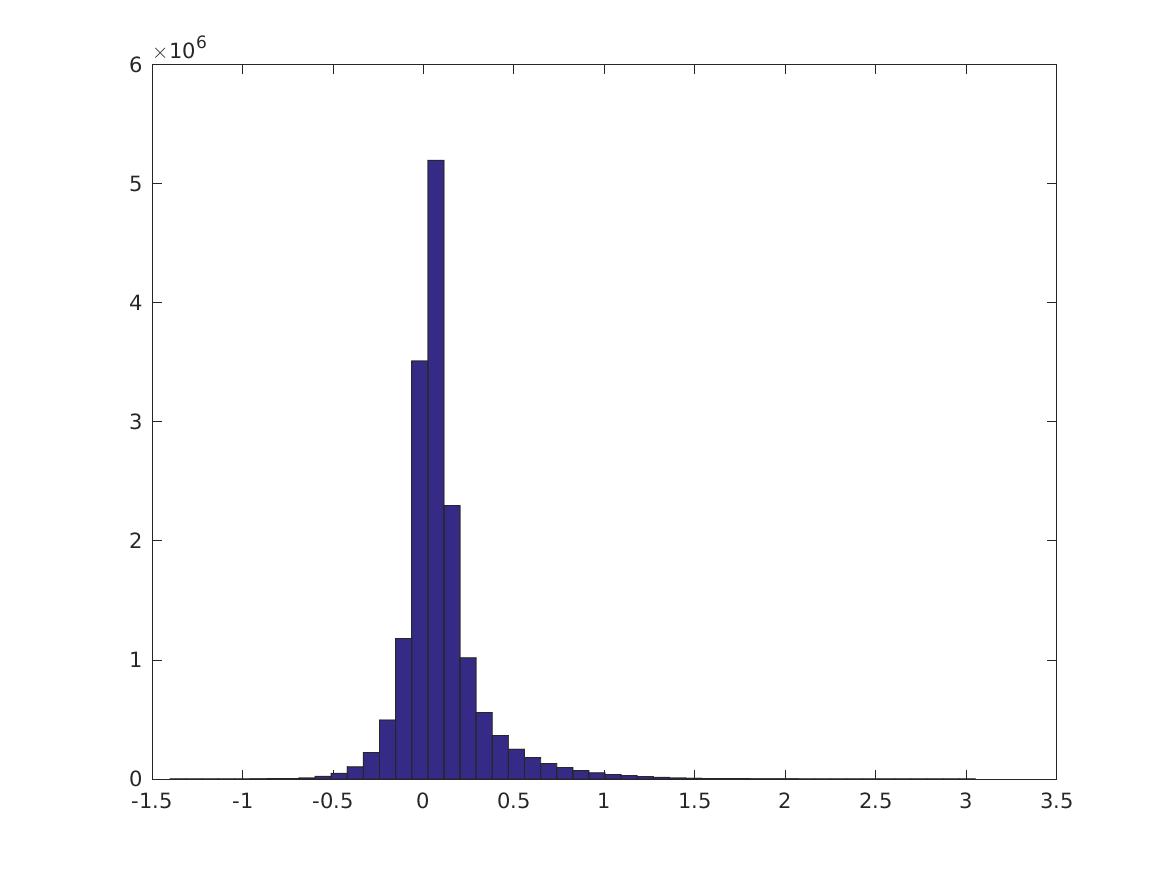

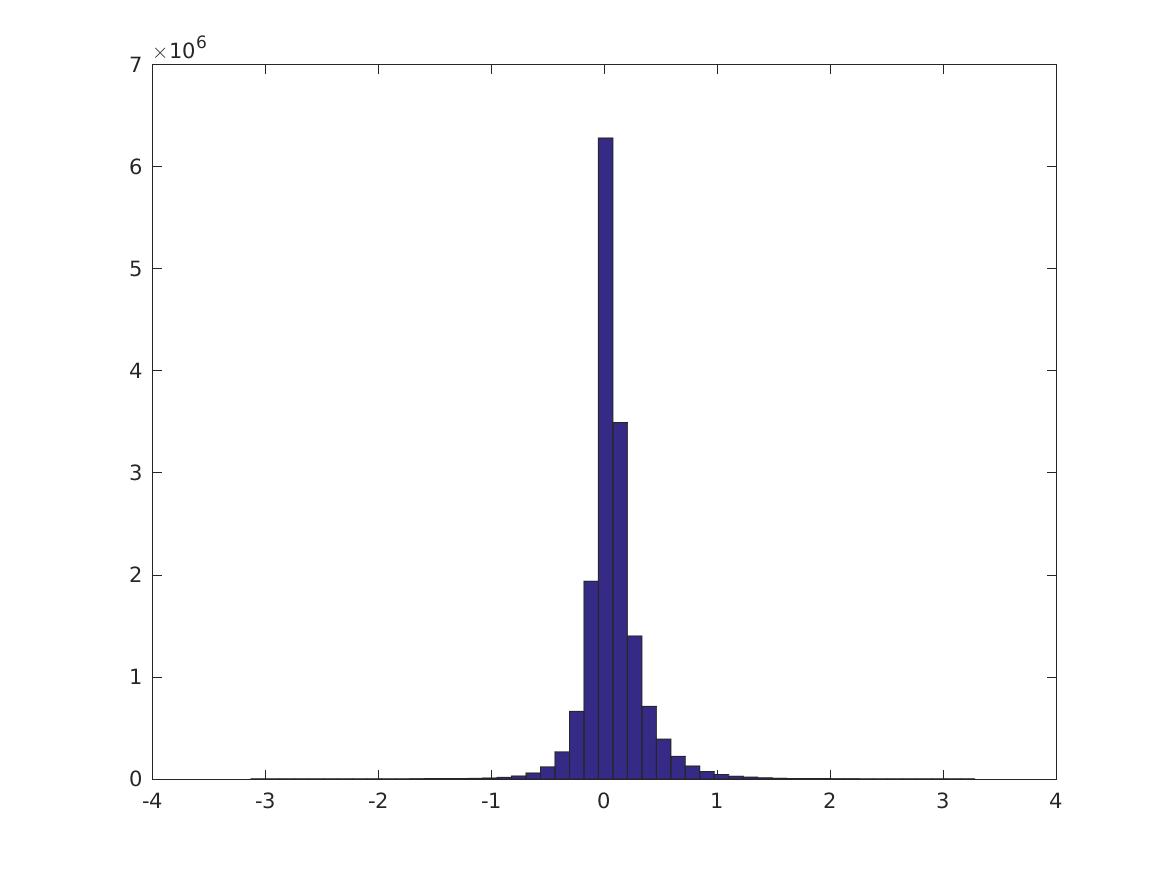

Here are the 0, 10,…, 100 percentiles of these distributions. The first 10 rows are the CPMI distributions for digits 0-9; the last row is the PMI distribution. It’s not always true that the CPMI’s for a digit are larger.
Columns 1 through 7
-1.3409 -0.0831 -0.0288 -0.0008 0.0178 0.0340 0.0520
-3.4897 -0.4067 -0.1484 -0.0196 0.0449 0.1054 0.1912
-1.2622 -0.1009 -0.0318 0.0013 0.0221 0.0404 0.0618
-1.3727 -0.0931 -0.0249 0.0067 0.0265 0.0458 0.0705
-2.5869 -0.0910 -0.0227 0.0091 0.0304 0.0524 0.0804
-1.4005 -0.0930 -0.0239 0.0113 0.0353 0.0582 0.0857
-2.6503 -0.0855 -0.0157 0.0134 0.0334 0.0555 0.0847
-3.1251 -0.1389 -0.0498 -0.0046 0.0237 0.0488 0.0796
-1.4074 -0.0984 -0.0277 0.0047 0.0240 0.0436 0.0701
-2.7278 -0.0846 -0.0149 0.0156 0.0372 0.0624 0.0966
-1.1984 -0.0727 -0.0094 0.0253 0.0534 0.0847 0.1241
Columns 8 through 11
0.0754 0.1111 0.1828 2.9385
0.3236 0.5373 0.9549 5.9331
0.0911 0.1378 0.2326 1.9094
0.1069 0.1677 0.2931 1.8452
0.1201 0.1838 0.3111 3.0054
0.1236 0.1841 0.3266 3.0518
0.1286 0.2034 0.3556 3.7582
0.1237 0.1959 0.3409 3.2767
0.1110 0.1844 0.3448 2.7204
0.1484 0.2350 0.4080 3.4744
0.1778 0.2587 0.4034 2.6897CODE: Run top_pmis_script which runs pmi_threshold_f and check_top_pmis_f.
Remove all features that do not have activation \(>1\) in some image. Now do the same thing. (NOTE: the maximum PMI is less because the max PMI’s are between features that have small maximum activation. Try this again thresholding by PMI?)
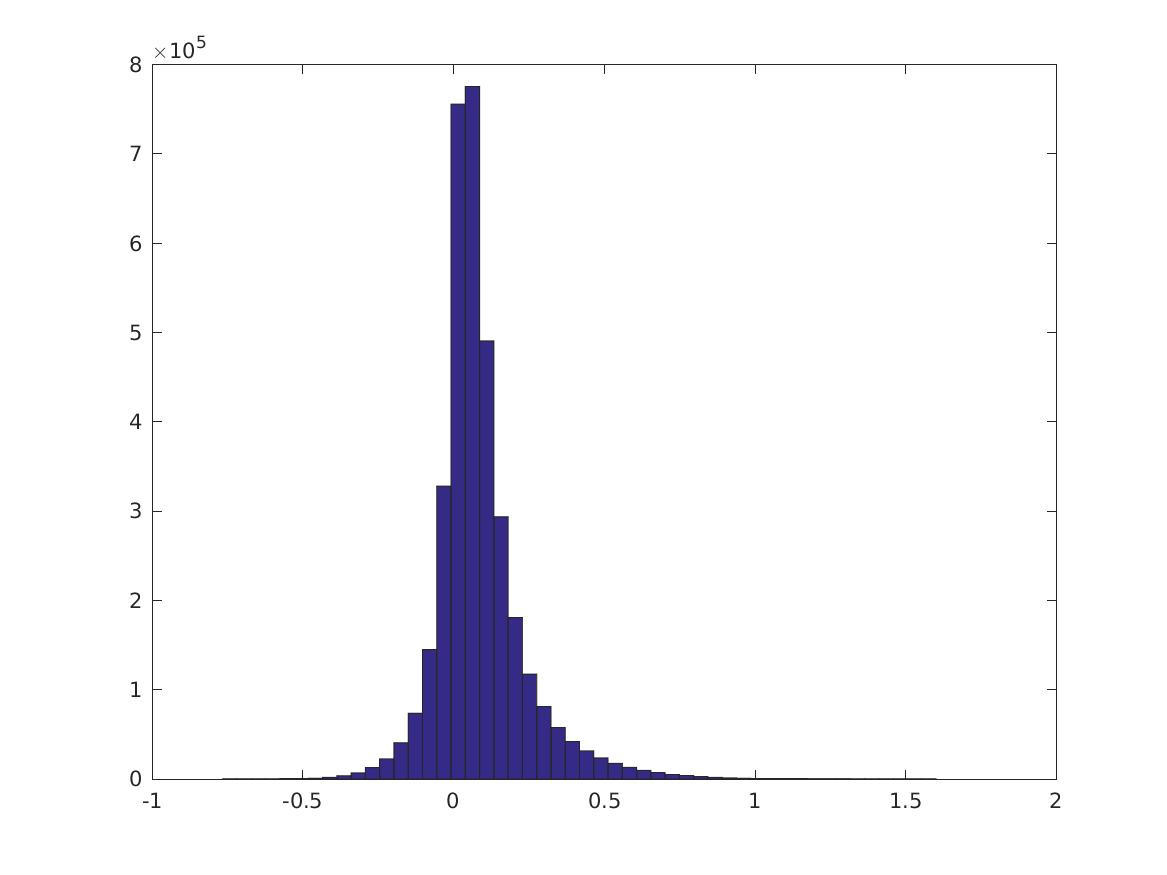
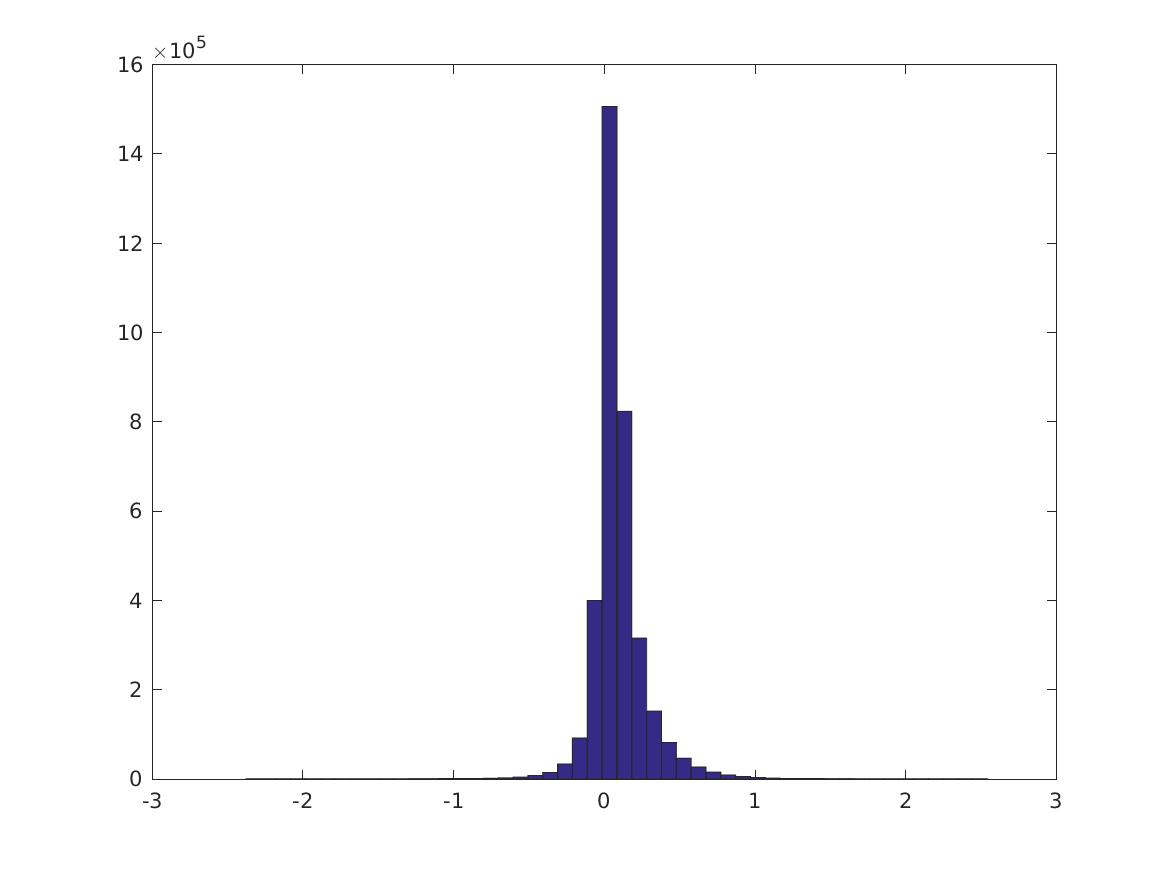
This is the distribution of \(PMI(v_i,v_j)\):

These are digits 0-9.








Percentiles of the distributions (\(i\)th row is 0, 10, 20, … 100% for the CPMI’s of digit \(i\), last row is PMI over all digits). (Note the last column is different)
Columns 1 through 7
-0.9746 -0.0588 -0.0140 0.0112 0.0306 0.0488 0.0681
-3.1365 -0.3102 -0.0940 0.0050 0.0609 0.1144 0.1812
-0.9300 -0.0626 -0.0130 0.0134 0.0333 0.0526 0.0740
-0.7660 -0.0462 -0.0022 0.0210 0.0403 0.0602 0.0835
-2.0740 -0.0565 -0.0071 0.0188 0.0397 0.0611 0.0855
-0.8674 -0.0628 -0.0077 0.0229 0.0471 0.0705 0.0966
-2.3770 -0.0457 -0.0007 0.0228 0.0428 0.0639 0.0886
-2.4824 -0.0928 -0.0235 0.0121 0.0379 0.0633 0.0925
-0.9435 -0.0521 -0.0066 0.0161 0.0347 0.0545 0.0785
-2.1549 -0.0505 -0.0020 0.0231 0.0446 0.0679 0.0961
-0.8693 -0.0515 0.0008 0.0344 0.0640 0.0949 0.1309
Columns 8 through 11
0.0910 0.1215 0.1730 1.5181
0.2772 0.4314 0.7470 5.3609
0.1007 0.1393 0.2117 1.5297
0.1141 0.1604 0.2521 1.6011
0.1169 0.1635 0.2537 2.2486
0.1293 0.1773 0.2773 2.0363
0.1215 0.1727 0.2771 2.5443
0.1305 0.1882 0.3019 2.9006
0.1111 0.1639 0.2788 2.1102
0.1344 0.1957 0.3210 2.7701
0.1761 0.2406 0.3557 1.7908
check_top_pmis produces a graph showing which features are close to each other. It’s not very enlightening.
CODE: Run pmi_suitability.m.
Is PMI the right measure? Are the feature vectors close to sparse? if the average image there is \(>20\%\) activation, then no.
For the first 10 images, this is what the percentiles look like:
percentiles:
pcs =
100 99 95 90 80 70
Image 1 (label 5)
2.3230
1.2966
0.8840
0.6766
0.4381
0.3140
Image 2 (label 0)
2.1275
1.3604
0.9482
0.7418
0.5036
0.3769
Image 3 (label 4)
1.7975
0.9874
0.6185
0.4704
0.3145
0.2247
Image 4 (label 1)
2.2506
1.2399
0.7160
0.4949
0.3029
0.1992
Image 5 (label 9)
2.0837
1.3970
0.8726
0.6461
0.3937
0.2707
Image 6 (label 2)
2.4831
1.4978
1.0466
0.7880
0.5002
0.3544
Image 7 (label 1)
2.5064
1.3688
0.7591
0.5203
0.3089
0.1935
Image 8 (label 3)
3.3030
1.6943
1.1588
0.8658
0.5756
0.4070
Image 9 (label 1)
2.2263
0.9396
0.4905
0.3279
0.1944
0.1178
Image 10 (label 4)
2.2420
1.3486
0.8494
0.6169
0.3751
0.2586
This looks pretty good—the top 1% of features are much larger than the rest.
Look for discriminative features, those that occur with one label and rarely for others. (I arbitrarily define this as: the maximum average activation is \(>1.5\) times the second largest, and the maximum is at least 0.1. I exclude the digit 1 (indexed as 2).)
sig_act_inds = intersect(find(m1>=1.5*m2), find(m1>0.1));
sig_act_inds2 = intersect(sig_act_inds, find(am1 ~=2));2080/7200 features fit the first criteria, 115/7200 of them are not 1. The distribution across labels is
0
1925
0
0
0
115
18
0
0
22
Some labels are not represented!
Do SVM coefficients have negative weights?
They have both positive and negative weights.
>> size(find(coeffs>0))
ans =
1 26333
>> size(find(coeffs<0))
ans =
1 30187CODE: Run svd_testing2.m, or sbatch slurm_svd.cmd. The results are saved in accs_1.mat.
Results:
| Dimension | Last | Best |
|---|---|---|
| Original | 99.34 | 99.44 |
| 500 | 99.26 | 99.44 |
| 100 | 97.5 | 98.82 |
| 50 | 96.84 | 96.84 |
Do weighted SVD (as in the NLP paper) and then train a SVM. Note this does worse than just SVD! Why?
| Dimension | Last | Best |
|---|---|---|
| 50 | 95.44 | 96.8 |
CODE: Run local_svd_svm.m and local_wsvm_script1.m.
Four settings:
(Dimension \(36\times 12 = 432\) total.)
| Type | Last | Best |
|---|---|---|
| SVD, separately | 99.18 | 99.24 |
| SVD, collected | 99.34 | 99.34 |
| WSVD, separately | 99.1 | 98.7 |
| WSVD, collected | 99.2 | 99.2 |
The training is stored in ../data/local_SVD.mat (got overridden, unfortunately) and ../data/local_wsvd_collected.mat.
One instance of training collected SVD.
accVal =
Columns 1 through 7
85.6800 85.6800 85.6800 85.6800 85.7200 85.9200 85.9400
Columns 8 through 14
86.3400 87.1600 87.9600 89.3800 91.2400 92.6200 94.1400
Columns 15 through 21
95.3400 96.1400 96.8600 97.3200 97.8400 98.1200 98.4800
Columns 22 through 28
98.5800 98.8400 98.8600 98.9200 98.9600 99.0600 99.1600
Columns 29 through 31
99.1800 99.3000 99.2000
WSVD, separate
Columns 1 through 7
92.1800 93.9200 95.0400 95.9400 96.6800 97.3400 97.8600
Columns 8 through 14
98.2200 98.4000 98.5800 98.7000 98.7600 98.9000 99.0000
Columns 15 through 21
99.0600 99.0400 99.0000 98.8600 99.1000 98.8800 98.3400
Columns 22 through 28
99.0000 98.9000 98.1600 98.9200 98.8200 99.0200 98.9800
Columns 29 through 31
98.9200 98.9600 98.7000
CODE: Run geo_pmi.m.
Hypothesis: The largest PMI values are for features that are for the same patch or adjacent patches.
The features are in a \(6\times 6\) grid; each of the 36 patches have a total of 200 features. We calculate the following PMI’s:
The collection process reduces the PMI’s. It seems that many of the largest PMI’s do occur in the same or adjacent patch.
collect, same patch; no collect, same patch; horiz collect; horiz no collect; vert collect; vert no collect; all
ans =
Columns 1 through 7
-0.9605 -0.3457 -0.2067 -0.1000 -0.0103 0.0783 0.1698
-1.0872 -0.0690 -0.0062 0.0281 0.0563 0.0880 0.1277
-1.5676 -0.3858 -0.2386 -0.1310 -0.0454 0.0409 0.1303
-0.9235 -0.0241 0.0307 0.0621 0.0919 0.1266 0.1695
-1.3763 -0.3810 -0.2237 -0.1053 -0.0072 0.0839 0.1708
-0.9163 -0.0108 0.0284 0.0530 0.0796 0.1093 0.1458
-1.1984 -0.0727 -0.0094 0.0253 0.0534 0.0847 0.1241
Columns 8 through 11
0.2735 0.3922 0.5678 1.3093
0.1821 0.2624 0.4080 2.4518
0.2220 0.3257 0.4557 1.1007
0.2240 0.3054 0.4534 2.6332
0.2607 0.3593 0.4863 0.9118
0.1942 0.2660 0.3970 2.6332
0.1778 0.2587 0.4034 2.6897No.
Pairs with large PMI aren’t necessarily adjacent. These are the top few PMI’s, with \((x_1,y_1), (x_2,y_2),\ve{(x_1,y_1)-(x_2,y_2)}\) (locations within the grid). (The following data is without thresholding and excluding 1’s; with thresholding it also looks similar.)
3.0000 4.0000 1.0000 1.0000 3.6056
1.0000 6.0000 1.0000 1.0000 5.0000
3.0000 5.0000 1.0000 1.0000 4.4721
2.0000 2.0000 1.0000 6.0000 4.1231
2.0000 2.0000 1.0000 1.0000 1.4142
1.0000 4.0000 1.0000 1.0000 3.0000
1.0000 1.0000 1.0000 1.0000 0
1.0000 4.0000 1.0000 1.0000 3.0000
3.0000 3.0000 1.0000 1.0000 2.8284
4.0000 5.0000 1.0000 1.0000 5.0000I sorted pairs of features by PMI value and took the largest 10000 pairs. The majority of the pairs (~77%) have strongest activation with the digit 1. (The digit 1 has conditional PMI values that are much larger than all the other digits - don’t know what to do about this.) 85% of the pairs have highest activations with the same digit.
If the pair has strong activation with 1, then it’s often much larger than the other activations.
Otherwise this is not necessarily true. A typical non-1 feature has average activations with the 10 digits that looks something like this: (NOTE: This was normalized—run again unnormalized to match the rest of the data.)
Columns 1 through 7
0.0708 0.2673 0.0940 0.1537 0.2122 0.1116 0.2627
Columns 8 through 10
0.2947 0.1448 0.3361